Lumencor’s ZIVA Light Engine for Yokogawa CSU-W1 featured at MBL’s Advanced Research Training Courses
The Marine Biological Laboratory (MBL) is a world-renowned leader in technical development and education in the field of light microscopy. Each year, MBL offers over 20 advanced research training courses in the biomedical sciences, providing participants with hands-on experience in various microscopy techniques using cutting-edge instrumentation.
One such technique, Spinning Disk Confocal (SDC) Microscopy, is widely used to study dynamic processes in living cells, tissues, or microorganisms through 3D imaging (1). The Yokogawa CSU-W1 Scanner is a highly regarded SDC scanner known for its remarkable scan speed, large field of view, and optimized light delivery. When paired with Lumencor’s ZIVA Light Engine, seven bright, stable, easy-to-implement lasers ensure intense, uniform illumination, facilitating user implementation and bolstering image quality. The ZIVA Light Engine offers a broader spectral range than traditional SDC lasers at a highly competitive price. As well, it is equipped with the essential filters and dichroics required to optimize spectral precision and contrast.
Throughout MBL’s courses, participants had the opportunity to use the ZIVA Light Engine with Yokogawa CSU-W1 on a variety of fixed and live specimens. Two Ph.D. students, Ella Nicklin and Zachary Sanchez, illustrated the use of the ZIVA 577 nm laser in imaging PCNA proteins in a Little Skate denticle and the exoskeleton of a marine copepod, respectively (Figure 1, 2). A third Ph.D. student, Bryce Dunn, explored the live cell imaging capabilities of the ZIVA Light Engine for Yokogawa CSU-W1 by imaging an Arabidopsis seedling expressing fluorescent cytoskeletal components using the CSU-W1 SoRa super resolution system (Figure 3).
Overall, users praised the integrated system for its simple operation, wide range of fluorescence excitation wavelengths, and exceptional quality of confocal and super resolution imaging. Below, read about a few user experiences with the ZIVA Light Engine for Yokogawa CSU-W1.
Ella Nicklin, Ph.D. Candidate in the Gareth Fraser Lab at University of Florida
“The quality and resolution of this spinning disk system are truly exceptional even without deconvolution. The clarity of the images it produces —especially with the silicon objective —really stands out. I particularly appreciate the 3D perspectives it offers, allowing us to optically slice through samples. This feature is especially beneficial for our thicker tissue sections.”
“Overall, the spinning disk confocal system has significantly enhanced our imaging quality and opened new possibilities for our research.”
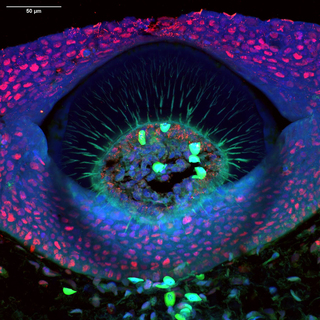
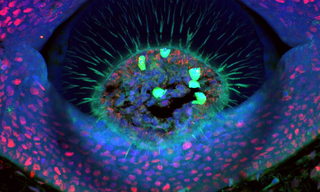
Figure 1. A cross-section through a denticle of a Little Skate (Leucoraja erinacea) embryo. DNA (blue) stained with DAPI, PCNA (red) with anti-PCNA primary and Alexa Fluor 594 secondary antibodies, and Collagen II (green) with anti-collagen II primary and Alexa Fluor 488 secondary antibodies. Autofluorescent blood cells are shown in green. Scale bar = 50 μm. Image was acquired using a ZIVA Light Engine for Yokogawa CSU-W1 Nikon Ti2 system with a 40x Silicon/1.25 NA objective.
Zachary Sanchez, Ph.D. Candidate in the Dylan Burnette Lab at Vanderbilt University
“It was incredible. It made everything identifiable. Everything was very clear.”
“The system with ZIVA was very easy to use. The pairing of equipment was high quality.”
Bryce Dunn, PhD Candidate at George Mason University
“You can clearly see components of the cell's cytoskeleton, which are often missing from textbook diagrams. The absence of signal in the cell's vacuole (on the right) is also a nice-looking feature of the image.”
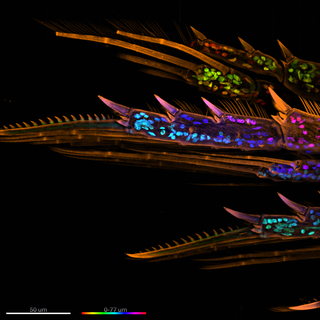
Figure 2. A maximum intensity projection of the legs of a marine Copepod at 60x magnification. The organism’s exoskeleton (orange) was stained with a WGA CF568 conjugate and DNA was stained with DAPI. The nuclei (DNA) are shown in rainbow colors to denote their z-plane location in the sample. Scale bar = 50 μm. Image was acquired using a ZIVA Light Engine for Yokogawa CSU-W1.
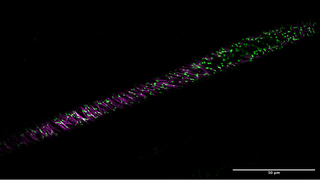
Figure 3. A cross section of a live Arabidopsis seedling at 20x magnification. The seedling expressed cytoskeleton components, mScarlet-Tubulin (magenta) and mNeonGreen-EB1 (green). Scale bar = 50 μm. Image was acquired using 488 nm (mNeonGreen) and 577 nm (mScarlet) excitation from the ZIVA Light Engine for Yokogawa CSU-W1.
- Oct 22, 2024