Background
High-content imaging of three-dimensional cellular model systems offers the potential for improving the clinical success rates of drug candidates by testing their activity under conditions that replicate in vivo tissue physiology and disease pathophysiology more accurately than two-dimensional cell cultures. Of course, three-dimensional specimens require three-dimensional imaging, so achieving this potential stretches the performance requirements for the principal components of the imaging system—the light source, microscope, and camera.
Needs
Dr. Tom Lummen, a microscopy engineer in the Department of Biosystems Science and Engineering, ETH Zürich University has kindly provided some insights into the requirements for implementing 3D live-cell imaging and how they are fulfilled using Lumencor’s CELESTA Light Engine. “I’m part of the microscopy team that operates the imaging core facility. We provide 18 high-end light microscopy workstations for the imaging needs of the users of our department. We are always scouting for new or expanded functionalities for these systems. We have a very broad array of imaging applications, these range from small subcellular features in live cells that have to be visualized on short timescales, all the way to the development of very big 3D organoids or tissue cultures that need to be monitored for weeks. Spinning disk confocal microscopy is becoming more in demand, as data acquisition needs are moving towards getting single-cell parameters but with very good statistics and/or time resolution. Our inverted spinning disk systems are geared towards live-cell observations for dynamic samples, but we have also seen an increase in demand for high 3D spatial throughput.”
Challenges
Spinning disk confocal microscopy allows for fast 3D imaging of live samples, but utilizing this technique involves striking a balance between competing demands. Dr. Lummen described some of the obstacles involved, and how they have affected him and his team, “Within microscopy there is always a trade-off when setting up a system, you can’t have all the maximum specs at the same time. What we try to go for are dedicated systems, but we try to keep them as tuneable as possible. We identified the need for a dedicated inverted spinning disk system that is geared towards high- throughput observation, whether spatial or temporal. This is when we started to explore what illumination technologies were out there. A suitable light source for high-throughput live-cell imaging at high speeds (to pair up with the speed of a spinning disk) would need to be highly flexible and deliver high intensity, temporally stable light output with the capability to rapidly switch between multiple spectral lines.”
Solution
Lumencor’s CELESTA Laser Light Engine was identified as an ideal illumination source for such dynamic live-cell spinning disk applications. Dr. Lummen shared his experiences with the CELESTA. “The CELESTA, in combination with the CrestOptics X-Light V3 spinning disk confocal scanner, Nikon Ti2 microscope and Teledyne Photometrics Kinetix sCMOS camera lets us image more cells in the same time span, which can proportionally increase the data throughput that we can offer to our users (Figure 1). The high intensity output of the CELESTA in combination with the large field of view (FOV) of the Kinetix camera allows us to image more cells per timepoint, or fewer tiles per composite large image. The combination with the large FOV of both the Nikon Ti2 and the CrestOptics X-Light V3 spinning disk confocal scanner let us configure a dual-camera large FOV setup (Figure 2). This enables us to further duplicate the data throughput or to perform true concurrent two-channel live-cell imaging in dynamic samples. Furthermore, the spectral output of the Lumencor CELESTA extends our imaging capabilities into the near-infrared (nIR).”
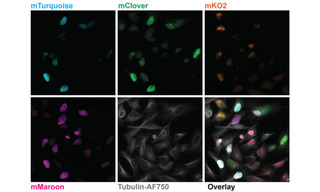
Figure 1. Five-color imaging on fixed Fucci4 HeLa cells. Images are sum intensity projections taken with a 100X, NA 1.45 oil immersion objective and a 25 mm FOV Kinetix camera. Recorded on a Nikon Ti2 microscope equipped with a CrestOptics X-light V3 spinning disk confocal scanner, a Lumencor CELESTA Light Engine, and a Teledyne Photometrics Kinetix sCMOS camera. Fucci4 is an indicator for cell cycle position based on cell cycle-dependent expression of the fluorescent proteins mTurquoise, mClover, mKO and mMaroon. Image credit: Javier Casares, D-BSSE, ETH Zurich.
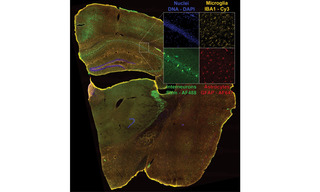
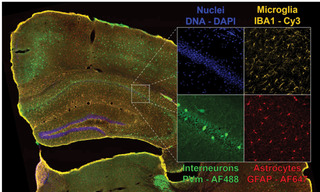
Figure 2. Four-color dual camera imaging of a fixed mouse brain slice. Image shows a maximum intensity projection composite of 8×9 acquisition tiles taken with a 20X NA 0.75 air objective with a 25 mm FOV Kinetix camera and a 15% tile overlap. Recorded on a Nikon Ti2 microscope equipped with a CrestOptics X-light V3 spinning disk confocal scanner, dual Teledyne Photometrics Kinetix sCMOS cameras, and a Lumencor CELESTA Light Engine.
- Jun 23, 2022