Case Study: Massively Multiplexed Single-Molecule Imaging Using Lumencor’s CELESTA Light Engine
MERFISH (multiplexed error-robust fluorescence in situ hybridization) is a massively multiplexed single-molecule imaging technology capable of simultaneously measuring the copy number and spatial distribution of hundreds to thousands of target RNA transcripts in single cells (Figure 1). In turn, single cell transcriptomic characterization allows in situ identification and spatial mapping of cell types in complex tissues. In MERFISH, identification of RNA targets is accomplished by hybridization of oligonucleotide probes carrying unique encoding sequences. The encoding sequences, and their associated RNA targets, are then identified by iterative optical interrogation. First, encoding oligonucleotide probes are hybridized to target RNAs. Then fluorescent dye-labeled readout oligonucleotide probes are successively added, spatially localized at the single molecule level and then extinguished [1,2]. Readout probes must hybridize exclusively to encoding probes, and not to cellular nucleic acids. The encoding probe identity is cumulatively deciphered across successive imaging cycles, with binary bit value “1” assigned if the encoding probe is detected by hybridization of a readout probe, and a “0” value if it is not. In principal, 12 hybridization+imaging cycles would be required to identify 4095 (212-1) unique encoding probes. In practice, the binary coding space is underpopulated to minimize false negative (1>0) and false positive (0>1) calling errors and consequent misidentification of target RNAs. For example, identification of 10,050 RNA targets using a 69-bit barcode has been demonstrated [3]. The number of hybridization+imaging cycles, and therefore the time required to complete data acquisition, can be reduced by simultaneous detection of readout probes with spectrally distinct fluorescent labels in each cycle (e.g. 23 imaging cycles x 3 fluorescent labels = 69 bits) [3].
MERFISH imaging requires a high intensity, spatially uniform illumination field matched to the dimensions of typical sCMOS camera sensors, (~200 mm2). To meet this requirement, a specialized critical epi-illuminator coupled to the fiber output from a CELESTA Light Engine (Lumencor, Inc., Beaverton, Oregon USA) is installed in the microscope, providing sufficient irradiance at the sample plane via a high magnification objective. The spectral output of the set of CELESTA Light Engine lasers, optimized for MERFISH multiplexed single-molecule imaging, are depicted in Figure 2. In typical MERFISH applications, the 477 nm, 637 nm and 748 nm laser outputs of the CELESTA Light Engine are used for detection of readout probes.
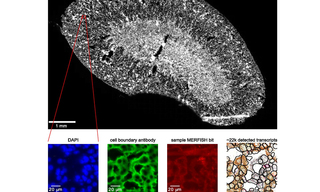
A sample multiplexed error-robust fluorescence in situ hybridization data acquisition workflow is depicted in Figure 1. The 405 nm CELESTA Light Engine laser is used for detection of DAPI and the 545 nm output is used for detection of fiducial markers used for spatial registration of adjacent fields of view. The DAPI channel for a mouse kidney tissue sample is represented in a low-resolution image. Red box indicates a zoomed-in region displayed in images B-E. DAPI (B) and (C) shows cell boundary antibody stain channels for the zoomed-in region. Panels B, C, and D show raw images with brightness and contrast levels selected for ease of visualization. Multiplexed error-robust fluorescence in situ hybridization signal for a single barcode bit channel in the same zoomed-in region is show in image E. There it’s possible to identify positions of decoded mRNA transcripts (colorful dots) and segmented cell boundaries (black) in the same zoomed-in region after running data through image analysis pipeline, with each color representing a unique gene species. Reproduced from [1] under CC BY 4.0. The high performance laser lighting CELESTA Light Engine provides, in terms of power and wavelength stability, spectral purity, and brightness, is critical to the precise imaging of such large, heterogeneous data samples for spatial biology with MERFISH.
![Figure 1. Sample multiplexed error-robust fluorescence in situ hybridization data acquisition workflow.(A) Low-resolution image of the DAPI channel for a mouse kidney tissue sample. Red box indicates zoomed-in region displayed in B-E. (B, C) DAPI (B) and (C) cell boundary antibody stain channels for the zoomed-in region. (D) Multiplexed error-robust fluorescence in situ hybridization signal for a single barcode bit channel in the same zoomed-in region. (E) Positions of decoded mRNA transcripts (colorful dots) and segmented cell boundaries (black) in the same zoomed-in region after running data through image analysis pipeline, with each color representing a unique gene species. Panels (B, C, D) show raw images with brightness and contrast levels selected for ease of visualization. Reproduced from [1] under CC BY 4.0](https://cms.lumencor.com/system/uploads/fae/image/asset/1003/xs_1.jpg)
Figure 1. Sample multiplexed error-robust fluorescence in situ hybridization data acquisition workflow.(A) Low-resolution image of the DAPI channel for a mouse kidney tissue sample. Red box indicates zoomed-in region displayed in B-E. (B, C) DAPI (B) and (C) cell boundary antibody stain channels for the zoomed-in region. (D) Multiplexed error-robust fluorescence in situ hybridization signal for a single barcode bit channel in the same zoomed-in region. (E) Positions of decoded mRNA transcripts (colorful dots) and segmented cell boundaries (black) in the same zoomed-in region after running data through image analysis pipeline, with each color representing a unique gene species. Panels (B, C, D) show raw images with brightness and contrast levels selected for ease of visualization. Reproduced from [1] under CC BY 4.0.
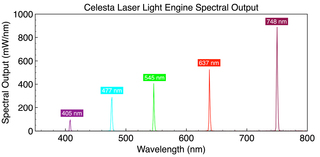
Figure 2. Spectral output of a CELESTA laser Light Engine optimized for MERFISH multiplexed single-molecule imaging.
- May 16, 2023
- (1) J Liu, V Tran, NF Neff et al., Concordance of MERFISH spatial transcriptomics with bulk and single-cell RNA sequencing. Life Sci Alliance (2022) 6:e202201701(opens in new window)
- (2) JR Moffitt, J Hao, X Zhuang et al., High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U S A (2016) 113:11046–11051(opens in new window)
- (3) C Xia, J Fan, G Emanuel, J Hao, X Zhuang et al., Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression Proc Natl Acad Sci U S A (2019) 116:19490–19499(opens in new window)


