Time-lapse Imaging with SOLA Light Engine
The Relevance of Time-lapse Imaging of GFP Expression Using the SOLA Light Engine to COVID-19 vaccine Efficacy
The delivery of mRNA through lipid-based transfection has been a longstanding challenge for the development of RNA therapeutics. Moreover, it has acquired a new and urgent prominence from the development of COVID-19 vaccines consisting of mRNAs encapsulated in lipid nanoparticles by Pfizer/BioNTech and Moderna. It is clearly important to understand the effects of mRNA-lipid complex formulation and extracellular medium composition on downstream expression of the protein immunogen that in turn determines vaccine efficacy. In 2019, before the start of the COVID-19 pandemic, a team of researchers from Ludwig-Maximilians-University in Munich and Stony Brook University, New York described the use of live-cell imaging on single-cell arrays (LISCA) to monitor the onset and rate of GFP expression following mRNA lipoplex transfection [1]. Single cells are arrayed on a micropatterned fibronectin substrate (Figure 1A), incubated with mRNA-lipid complexes for 1 hour and then monitored by time-lapse fluorescence microscopy for 20 hours (Figure 1B). For GFP fluorescence to give an authentic representation of protein expression levels, stable and reproducible excitation is essential, making the SOLA Light Engine the ideal high-performance illumination source for this application. As well as characterizing the pronounced cell-to-cell variability in onset times and rates of protein expression (Figure 1), LISCA was used to determine the effect of serum proteins on the cellular uptake of different mRNA-lipid complex formulations.
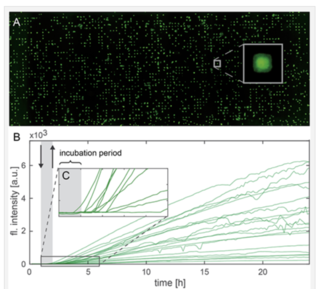
Figure 1: (A) Single GFP-expressing HuH7 cells arrayed on micropatterned fibronectin. (B) Single cell fluorescence trajectories representing GFP expression. The gray-shaded area represents the initial 1-hour period of incubation with mRNA-lipid complexes. (C) Enlarged region of (B) showing cell-to-cell variation in onset of protein expression. Reproduced from Reiser et al. (2019) [1] under the terms of the Creative Commons Attribution License.