Investigation into 3D spatial organization of chromatin, how precise interactions between genes and distal regulatory sequences exercise control over transcription, have advanced significantly through the application of techniques such as high-throughput chromosome conformation capture (Hi-C) and fluorescence in situ hybridization (FISH). While powerful, these methods have limitations such as low throughput capabilities, extensive sequencing requirements, or low spatial resolution, which hinder their ability to provide comprehensive information. Optical Reconstruction of Chromatin Architecture (ORCA), a microscopy-based approach that overcomes these limitations, utilizes high-resolution microscopy in conjunction with Oligopaint and RNA-FISH to visualize chromatin organization at single-cell resolution in both cryo-sectioned tissues and cultured cells (1). In ORCA imaging, primary oligonucleotide probes are designed with unique barcodes and are used to interrogate specific regions of chromatin. After imaging and analysis of the barcodes and determination of their positions relative to one another, researchers can construct a distance map to understand the structural organization of the chromatin region within the cell nucleus (Figure 1) (2). Developed by scientists in the laboratory of Alistair Boettiger at Stanford University, ORCA offers simultaneous identification of chromatin structural properties and information on individual genomic regions within single cells, comparable to Hi-C and FISH methods (1). After utilizing free-space coupled lasers in their prototype ORCA systems, the Boettiger lab recently upgraded one of their illumination systems to incorporate the CELESTA Light Engine (1,3). The CELESTA integrates an array of independent, solid-state lasers to provide stability, optical power, and brightness in a compact, turnkey illuminator with a space-saving footprint. Using the fiber-coupled CELESTA and large field-of-view optics for immunofluorescence and ORCA imaging, Boettiger and colleagues tested current models on the repression of Hox genes by Polycomb group proteins (3). Their findings provided new insights into how protein complexes organize chromatin and contribute to the silencing of target genes by preventing transcription.
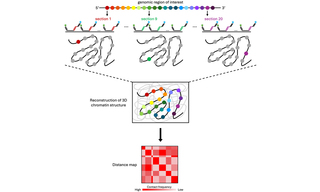
Figure 1. Optical reconstruction of chromatin architecture (ORCA) renders linear DNA sequences in their 3-dimensional cellular context. A set of primary oligonucleotide probes is designed to identify a genomic region of interest and partitions the region into short segments (colored circles). Each probe carries a unique barcode that is iteratively interrogated by sequential imaging steps to provide the 3D position of each probe/barcode in the nucleus. The 3D positions of barcodes are measured and presented as pseudocolors, leading to the 3D reconstruction of chromatin architecture in the regions of interest. The 3D positions between each pair of barcodes are used to construct a distance map.
- May 16, 2024
- (1) Visualizing DNA folding and RNA in embryos at single-cell resolution. LJ Mateo, SE Murphy, A Hafner, IS Cinquini, CA Walker, AN Boettiger (2019). Nature 568:49–54.(opens in new window)
- (2) Tracing DNA paths and RNA profiles in cultured cells and tissues with ORCA. LJ Mateo, N Sinnott-Armstrong, AN Boettiger (2021). Nat Protoc. 16:1647–1713.(opens in new window)
- (3) Polycomb repression of Hox genes involves spatial feedback but not domain compaction or phase transition. SE Murphy, AN Boettiger (2024). Nat Genet. 56:493–504.(opens in new window)


